Blair RM, Fang H, Branham WS, Hass BS, Dial SL, Moland CL, Tong W, Shi L, Perkins RG, and Sheehan DM. “The estrogen receptor relative binding affinities of 188 natural and xenochemicals: Structural diversity of ligands.” Toxicol Sci. 2000; 54:138-153.
Branham WS, Dial SL, Moland CL, Hass BS, Blair RM, Fang H, Shi L, Tong W, Perkins RG, and Sheehan DM. “Binding of phytoestrogens and mycoestrogens to the rat uterine estrogen receptor.” J Nutr. 2002; 134:658-664.
Cheng F, et al. J Chem Inf Model. 2012; 52(3):655-669.
Fang H, Tong W, Shi LM, Blair R, Perkins R, Branham W, Hass BS, Xie Q, Dial SL, Moland CL, and Sheehan DM. “Structure-activity relationships for a large diverse set of natural, synthetic, and environmental estrogens.” Chem Res Tox. 2001; 14:280-294.
Fang H, Tong W, Branham WS, Moland CL, Dial SL, Hong H, Xie Q, Perkins R, Owens W, Sheehan DM. “Study of 202 Natural, Synthetic, and Environmental Chemicals for Binding to the Androgen Receptor.” Chem Res Toxicol. 2003; 16: 1338-1358.
Hamelink JL. “Current bioconcentration test methods and theory.” in Aquatic Toxicology and Hazard Evaluation. Eds. Mayer FL and Hamelink JL. West Conshohocken, PA ASTM STP, 1977.
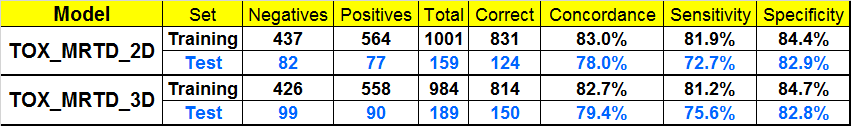
Matthews EJ, Kruhlak NL, Benz RD and Contrera JF “Assessment of the health effects of chemicals in humans: I. QSAR estimation of the maximum recommended therapeutic dose (MRTD) and no effect level (NOEL) of organic chemicals based on clinical trial data.” Current Drug Discovery Technologies 2004; 1:1
Schultz, TW, “TETRATOX: Tetrahymena pyriformis population growth impairment endpoint – A surrogate for fish lethality.” Toxicol Methods. 1997; 7:289-309; https://www.vet.utk.edu/TETRATOX/
Vinggaard AM, Niemelä J, Wedebye EB, Jensen GE. “Screening of 397 Chemicals and Development of a Quantitative Structure – Activity Relationship Model for Androgen Receptor Antagonism.” Chem Res Toxicol. 2008; 21: 813-823.
Zhu H, Tropsha A, Fourches D, Varnek A, Papa E, Gramatica P, Öberg T, Dao P, Cherkasov A, Tetko IV, “Combinatorial QSAR Modeling of Chemical Toxicants Tested Against Tetrahymena pyriformis.” J Chem Inf Model. 2008; 48:766-784.