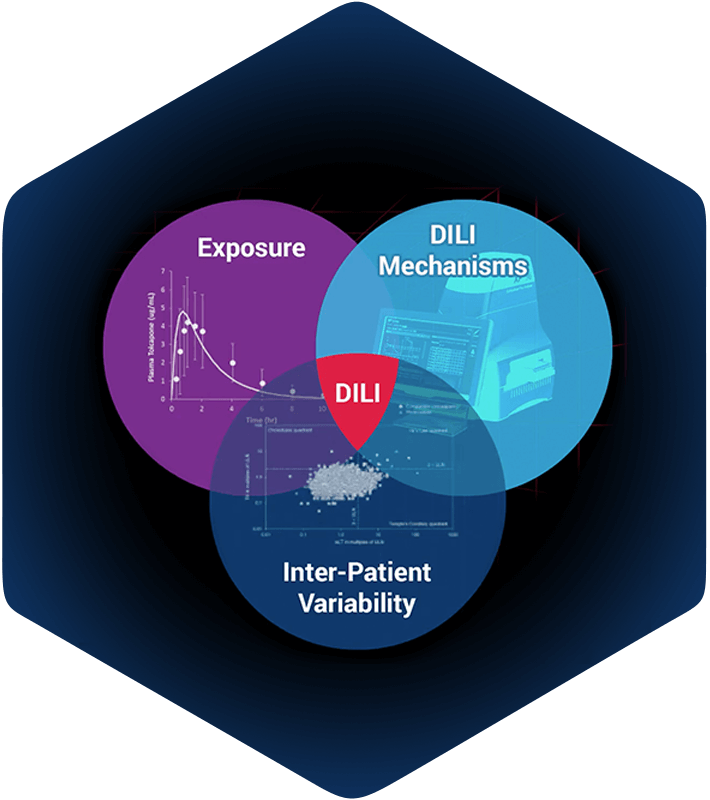
DILIsym simulations integrate compound exposure with compound mediated mechanisms of intrinsic drug toxicity, and with inter-individual variability. The intersection of these is prediction of hepatotoxicity for:
- Phase II, III, or IV clinical trials in patients
- First-in-human clinical trials in healthy volunteers
- Small preclinical species
- Large preclinical species
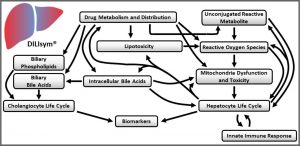
DILIsym is a “middle-out”, multi‑scale representation of drug-induced liver injury. It includes key liver cell populations (e.g., hepatocytes, Kupffer cells), bile duct cells (cholangiocytes), intracellular biochemical systems (e.g., mitochondrial function), and whole body dynamics (e.g., drug distribution and metabolism). DILIsym represents these biochemical and physiological processes for mice, rats, dogs, and humans, based on a substantial amount of publicly-available literature. Through the generation of SimPops® (a collection of simulated patients forming a simulated population), DILIsym also includes inter‑individual variability.
DILIsym includes a graphical user interface (GUI) to facilitate in silico investigations. The GUI permits the specification of new simulated experiments, including compound, dose, dosing frequency, duration, mechanism(s), and species. Simulations can be performed on individuals or populations. Simulation results can be visualized within the GUI or exported for further analysis in other software. Simulations can also be run with the command-line version of DILIsym, making integration into a stream-lined workflow possible. A high performance grid license (HPGL) option is available via licensing (or included with DILI-sim Initiative membership), which allows for use of an unlimited number of command-line-based simulation engines and enables users to scale up and complete large batches of simulations on internal or external compute clusters.
DILIsym prospectively supports key management decisions by providing information about potential drug-induced liver injury risk in upcoming experimental or clinical study designs, as well as mechanistic rationale for the underlying biochemical events that could cause liver toxicity. DILIsym also supports key management decisions about compounds for which liver transaminase elevations have already been observed by identifying patient screening or dosing protocols to mitigate risk. Additionally, DILIsym can mechanistically interpret existing data on liver transaminase elevations to predict the underlying level of hepatocyte loss and rate of liver recovery, supporting the distinction of innocuous vs. dangerous liver signals.